Cell cycle and cell cycle regulation: The cell cycle is divided into three stages: interphase, mitosis, and cytokinesis. Interphase is the period between successive rounds of nuclear division and is characterized by cell growth and synthesis of new DNA. Interphase is further divided into G1, S, and G2 phases. Division of genetic material occurs during mitosis, which is divided into prophase, prometaphase, metaphase, anaphase, and telophase. Cytokinesis is the division of the cytoplasmic material.
Following are the phases of interphase -
i) G1 and G0: G0 (resting phase) is seen when the cell is in a quiescent state. Cells in G0 are not dividing. Permanent cells remain in G0, while other cells can transition from G0 to G1 when they re-enter the cell cycle to undergo cell division. During G1, the cell grows; RNA and protein synthesis occur; and organelles and intracellular structures are duplicated. The length of the G1 phase differs between cells: it is shorter in rapidly dividing cells and longer in slowly dividing cells.
ii) S phase: Synthesis of nuclear DNA (DNA replication) occurs during S phase. The cell replicates its DNA content and goes from 2N to 4N by the end of S phase. Here, N stands for each copy of a chromosome, so 2N means 2 copies of the same chromosome. Haploid is 1N, while diploid is 2N.
iii) G2 phase: This is the phase between S phase and mitosis.
Following phases are seen in mitosis -
i) Prophase: Genetic material condenses into chromatids, with each chromatid connected to its sister chromatid at a centromere. Protein complexes called kinetochores form and attach to the chromatids. The mitotic spindle forms, and the nucleolus disassembles.
ii) Prometaphase: The nuclear envelope disassembles. Microtubules of the mitotic spindle attach to kinetochores, allowing the chromosomes to be pulled apart.
iii) Metaphase: Chromosomes align at the equator of the mitotic spindle.
iv) Anaphase: The centromere splits, sister chromatids separate, and they move to opposite poles of the mitotic spindle. Kinesins push apart the spindle to elongate it.
v) Telophase: The mitotic spindle disassembles. A nuclear envelope forms around each set of chromatids at the poles, chromatids decondense, and nucleoli form again in the daughter nuclei. Two daughter nuclei are formed from each nucleus.
Following events happen in cytokinesis -
i) An actin microfilament ring forms and contracts to produce a cleavage furrow.
ii) The furrow deepens and separates the cell into two daughter cells.
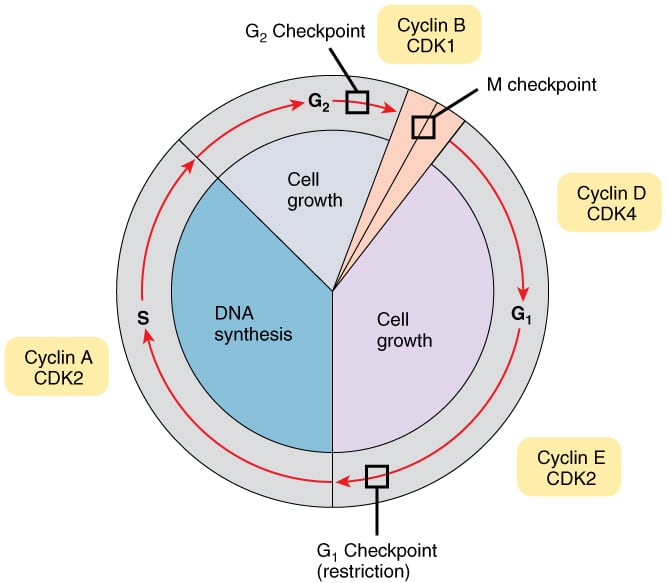
Regulation of cell cycle: The timing of events in the cell cycle is orchestrated by the expression of cyclins. Cyclins bind and activate CDKs (cyclin-dependent kinases), which phosphorylate several structural proteins and enzymes, triggering different cell cycle events.
There are four classes of cyclins that are expressed during different stages of the cell cycle - cyclin D (G1), cyclin E (G1/S), cyclin A (S), and cyclin B (mitosis). Following CDKs are expressed in different stages of the cell cycle - CDK4 and CDK6 (G1), CDK2 (G1/S and S), CDK1 (mitosis).
Gene expression of a cyclin is turned on just before its stage of the cell cycle. Once that stage is complete, the cyclin is ubiquitinated and targeted for degradation by the proteasome. Cyclins activate CDKs by inducing a conformational change in CDKs that opens an ATP-binding pocket. CDK activity is also regulated by phosphorylation.
Cyclin D turns on the expression of G1/S and S cyclins. DNA damage also inhibits the cell cycle by inhibiting the activation of G1/S-CDK complexes. Expression of G1/S cyclins is regulated by E2F proteins, which are transcription activators. Rb proteins bind to and inhibit the activity of E2F1, E2F2, and E2F3. Other Rb family proteins, p107 and p130, function as corepressors with E2F4 and E2F5. Mitogens stimulate cell division by increasing the amount of G1 cyclins.
Ink4 proteins bind to cyclin D-CDK and inhibit its activity. Expression of Ink4 is increased in the presence of the transcription protein Miz1, which in turn is activated by TGF beta. Mitogens lower the expression of Ink4 proteins.
TOR (target of rapamycin), a serine/threonine signalling pathway, is considered the master regulator of growth rates in cells because it increases the expression of ribosomal proteins and RNAs and increases the rate of translation. Growth factors like IGF 1 and hormones stimulate the activity of TOR.
Checkpoints monitor the progress of cell cycle events and prevent entry into subsequent stages until the current stage is completed. Three checkpoints monitor the progress of the cell cycle and regulate the activity of cyclin-CDK complexes.
i) G1 checkpoint: The restriction point is seen in G1 phase. Any cell that passes this checkpoint will continue with cell division. In the presence of DNA damage, tumor suppressor proteins halt the cell cycle before it reaches the restriction point. Mutations in tumor suppressor genes allow a cell with damaged DNA to divide, which can cause cancer. The G1 checkpoint also ensures that cells do not enter S phase before completion of G1.
ii) S phase checkpoint: In the presence of DNA damage in S phase, DNA synthesis and the cell cycle are slowed. BRCA 1 plays a role in the repair of double-stranded DNA breaks in S phase.
iii) G2 checkpoint: This checkpoint occurs before transitioning from G2 to M phase. CDK1 is inactivated by phosphorylation throughout G1 and S phases. In G2, CDK1 is dephosphorylated by cdc25C phosphatase, which allows it to bind to and activate cyclin B so that the cell can move forward into mitosis. Tumor suppressors ATM and ATR inhibit cdc25C phosphatase and halt the cell in G2 when needed.
Sign up for free to take 2 quiz questions on this topic